Create Workflow
For this tutorial, the user will build a simple QC Workflow that records a QC value and calculates a Pass/Fail result as a Report. By this point, you’ve already seen this Workflow in other tutorials, so we’ll focus on how to create the content in the context of the L7|ESP SDK.
For this demo, our Workflow will have two parts:
A Protocol to capture QC metadata about a Sample.
A Protocol to generate a Report using a Pipeline.
Here’s a schematic detailing the components of that Protocol at a high level:
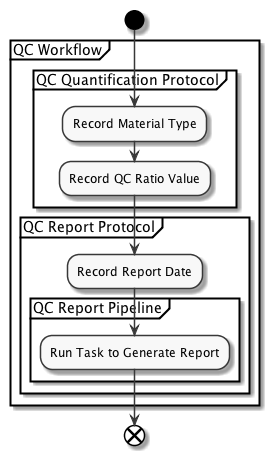
All content developed within the L7|ESP SDK is defined within YAML config files that live in the content directory within the repository. For more information about the structure of the repository, see the Project Structure section of the documentation.
Step 1: Create a Protocol
Now that we know what type of content we’re going to create, let’s make some content files. First, we need to create our first Protocol (QC Quantification):
Using the L7|ESP SDK, we can create this configuration via YAML config file. For more information on the YAML config format or types of content that can be created with the YAML config format, please see the L7|ESP Python client documentation.
For this first Protocol, create the following file:
# Contents of: content/protocols/QC-Quantification.yml
QC Quantification:
protocol: standard
variables:
- Type:
rule: dropdown
dropdown:
- 'DNA'
- 'RNA'
- Ratio:
rule: numeric
In this file, we’ve defined an L7|ESP Protocol that has two columns the user will input when filling out Worksheets: Type and Ratio. Type is a column of dropdown type with options DNA and RNA, and Ratio is a numeric column. Other types of Protocol columns include (also listed in the L7|ESP Python client documentation):
string- Column with text or string data.
numeric- Column with numeric data.
dropdown- Column with a dropdown selector. The options for the dropdown are specified as a list in thedropdownproperty.
checkbox- Column with checkbox indicating yes/no status.
date- Column with date picker for selecting calendar date.
attachment- Column for uploading file attachment.
link- Column with data containing external link.
barcode- Column for scanning barcode data and printing various types of barcodes (QR, 1D, Mini DataMatrix, etc.).
Note
This list may not be fully comprehensive. For the full list, please see the L7|ESP Python client documentation.
Along with this Protocol, we’re going to define an L7|ESP Pipeline and pipeline Protocol to include in our Workflow.
Step 2: Create a Pipeline to generate a Report:
To create a Pipeline for use in a Workflow, we need to create two things:
A Pipeline that will generate a Report when given Type and Ratio QC data.
A pipeline Protocol that will link the values from the QC Quantification Protocol to our Pipeline.
To start, define the report-generating Pipeline:
# Contents of: content/pipelines/QC-Report-Pipeline.yml
QC Report Pipeline:
report:
name: Result Report
elements:
- - depends:
- {file: Result Report, tasknumber: 1}
type: raw_file
- []
tasks:
- Generate QC Report:
desc: Task to generate QC report from workflow metadata
cmd: |+
# Simple Task that determines if the specified 'ratio' is in the proper range
RNA_MIN=1.8
RNA_MAX=2.1
DNA_MIN=1.7
DNA_MAX=2.0
TYPE="{{ material }}"
RATIO="{{ ratio }}"
if [ $TYPE = "RNA" ]; then
PASS=`echo "$RNA_MIN <= $RATIO && $RATIO <= $RNA_MAX" | bc`;
elif [ $TYPE = 'DNA' ]; then
PASS=`echo "$DNA_MIN <= $RATIO && $RATIO <= $DNA_MAX" | bc`;
fi
echo $PASS
if [[ $PASS = 1 ]]; then
echo "<b>Your sample <font color='green'>Passed</font> and contains pure $TYPE</b>" >> result.html
else
echo "<b>Your sample <font color='red'>Failed</font> and is NOT pure $TYPE</b>" >> result.html
fi
files:
- Result Report:
file_type: html
filename_template: "{{ 'result.html' }}"
In the block above, we’re defining a Pipeline with a single Task. In that Task, the cmd block contains code that will be run as part of the Pipeline. This code can either be bash code or a call to an external script that will run the Pipeline. Also in the cmd block are references to {{ material }} and {{ ratio }} variables that are passed into the Pipeline. Next, we’ll create configuration for a Protocol that can run this Pipeline as part of our Workflow.
Step 3: Create a ‘pipeline’ Protocol to record data and run the Pipeline:
Now, let’s create our pipeline Protocol for running the Pipeline that we defined above. For this Protocol, we want to capture column data (Type and Ratio) from a previous Protocol so they can be passed as arguments (material and ratio) to the Pipeline. We also want to include a date column for users to specify a date when the QC data were taken.
Create the following file to capture all of this information:
# Contents of: content/protocols/QC-Report.yml
QC Report:
protocol: pipeline
pipeline: QC Report Pipeline
variables:
- material:
rule: text
value: "{{ column_value('Type', 'QC Quantification') }}"
visible: false
- ratio:
rule: numeric
value: "{{ column_value('Ratio', 'QC Quantification') }}"
visible: false
- Report Date:
rule: date
Above, we can reference our Pipeline using the pipeline configuration option. Additionally, for Protocols that are executing a Pipeline, protocol: pipeline must be explicitly specified as part of the Protocol definition. Finally, to carry data over from a previous Protocol, we’re using an L7|ESP expression in the value field for the column. For more information on expressions and how they’re used in L7|ESP, see the L7|ESP documentation.
Step 4: Put it all together in a Workflow:
Now that we our Pipeline and Protocols defined, we can put them all together in a Workflow that can be used in the lab.
Define the Workflow as follows:
# Contents of: content/workflows/QC-Workflow.yml
QC Workflow:
desc: Workflow to measure QC values and generate a QC report.
tags:
- quickstart
- qc
protocols:
- QC Quantification
- QC Report
Above, since our Protocol configuration is already defined, we can simply reference them in the protocols block of the Workflow.
Notes
You can also define all of these items in a nested way.
For instance, if you want to define the QC Workflow with a single Protocol all in the same config file, instead, define the Workflow this way:
# workflow
QC Workflow:
desc: Workflow to measure QC values and generate a QC report.
tags:
- quickstart
- qc
# nested protocols
protocols:
- QC Quantification:
protocol: pipeline
variables:
- Type:
rule: dropdown
dropdown:
- 'DNA'
- 'RNA'
- Ratio:
rule: numeric